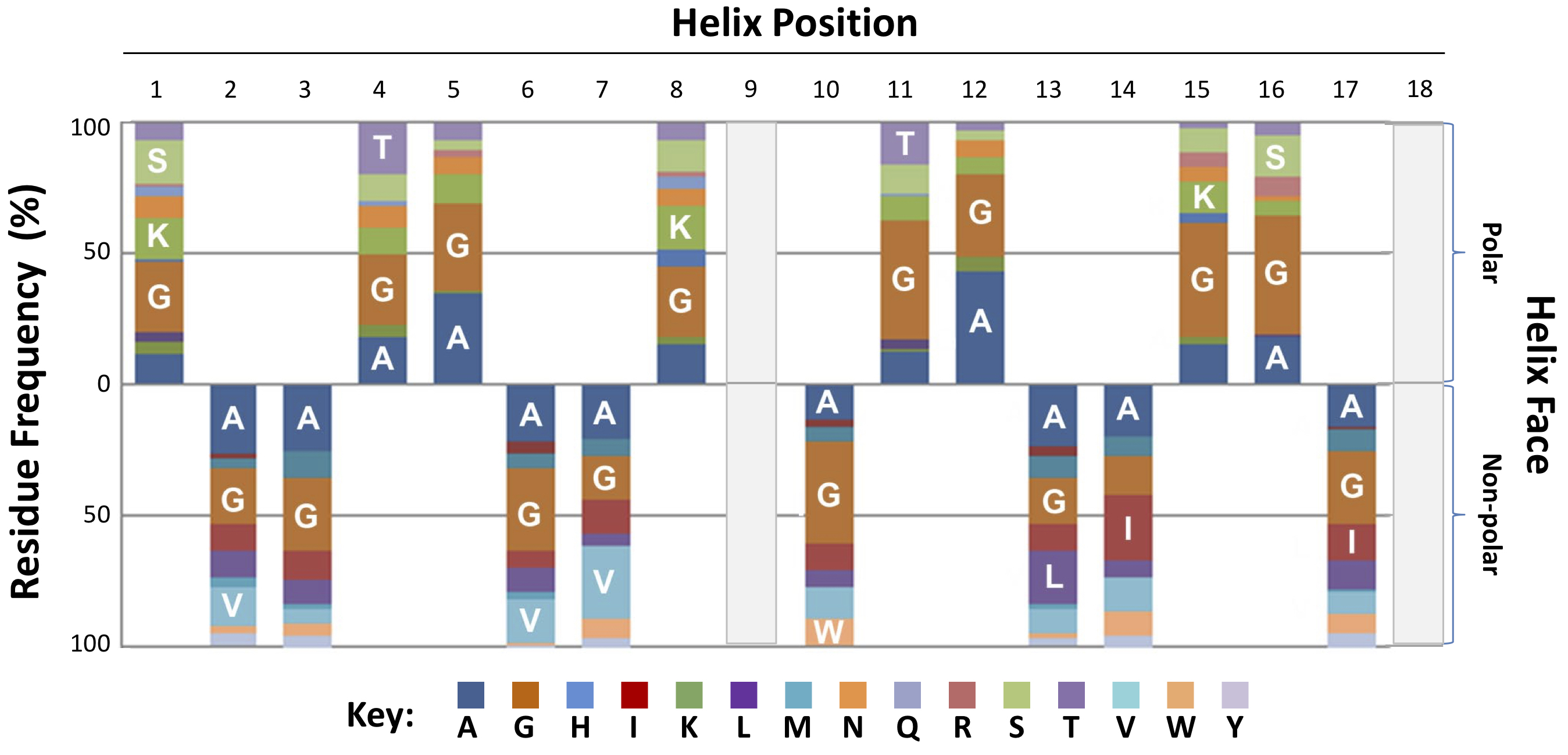
 Sequence formula for Type II bacteriocins based on 3D spatial localization of residues
Sequence formula for Type II bacteriocins based on 3D spatial localization of residues
Discovery of Novel Type II Bacteriocins Using a New High-Dimensional Bioinformatic Algorithm
Abstract
Antimicrobial compounds first arose in prokaryotes by necessity for competitive self-defense. In this light, prokaryotes invented the first host defense peptides. Among the most well-characterized of these peptides are class II bacteriocins, ribosomally- synthesized polypeptides produced chiefly by Gram-positive bacteria. In the current study, a tensor search protocol—the BACII-alpha algorithm—was created to identify and classify bacteriocin sequences with high fidelity. The BACII-alpha algorithm integrates a consensus signature sequence, physicochemical and genomic pattern elements within a high-dimensional query tool to select for bacteriocin-like peptides. It accurately retrieved and distinguished virtually all families of known class II bacteriocins, with an 86% specificity. Further, the algorithm retrieved a large set of unforeseen, putative bacteriocin peptide sequences. A recently-developed machine-learning classifier predicted the vast majority of retrieved sequences to induce negative Gaussian curvature (NGC) in target membranes, a hallmark of antimicrobial activity. Prototypic bacteriocin candidate sequences were synthesized and demonstrated potent antimicrobial efficacy in vitro against a broad spectrum of human pathogens. Therefore, the BACII-alpha algorithm expands the scope of prokaryotic host defense bacteriocins and enables an innovative bioinformatics discovery strategy. Understanding how prokaryotes have protected themselves against microbial threats over eons of time holds promise to discover novel anti-infective strategies to meet the challenge of modern antibiotic resistance.