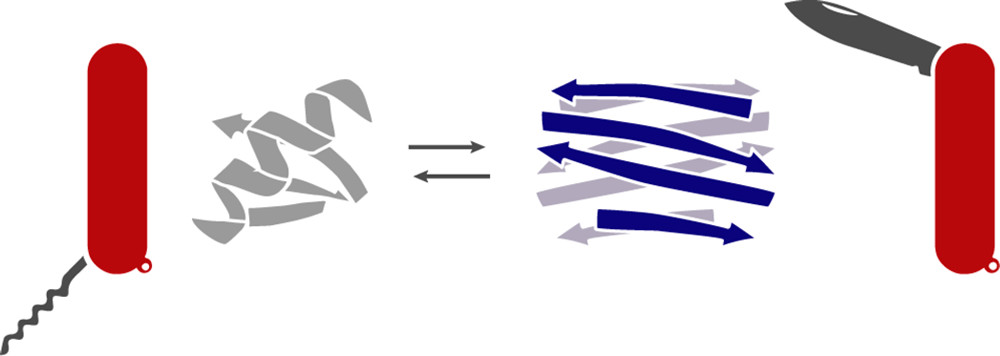
 XCL1 is a metamorphic chemokine that can toggle between a monomer and dimer structure
XCL1 is a metamorphic chemokine that can toggle between a monomer and dimer structure
Switchable membrane remodeling and antifungal defense by metamorphic chemokine XCL1
Abstract
Antimicrobial peptides (AMPs) are a class of molecules which generally kill pathogens via preferential cell membrane disruption. Chemokines are a family of signaling proteins that direct immune cell migration and share a conserved α−β tertiary structure. Recently, it was found that a subset of chemokines can also function as AMPs, including CCL20, CXCL4, and XCL1. It is therefore surprising that machine learning based analysis predicts that CCL20 and CXCL4’s α-helices are membrane disruptive, while XCL1’s helix is not. XCL1, however, is the only chemokine known to be a metamorphic protein which can interconvert reversibly between two distinct native structures (a β-sheet dimer and the α−β chemokine structure). Here, we investigate XCL1’s antimicrobial mechanism of action with a focus on the role of metamorphic folding. We demonstrate that XCL1 is a molecular “Swiss army knife” that can refold into different structures for distinct context-dependent functions: whereas the α−β chemokine structure controls cell migration by binding to G- Protein Coupled Receptors (GPCRs), we find using small angle X-ray scattering (SAXS) that only the β-sheet and unfolded XCL1 structures can induce negative Gaussian curvature (NGC) in membranes, the type of curvature topologically required for membrane permeation. Moreover, the membrane remodeling activity of XCL1’s β-sheet structure is strongly dependent on membrane composition: XCL1 selectively remodels bacterial model membranes but not mammalian model membranes. Interestingly, XCL1 also permeates fungal model membranes and exhibits anti-Candida activity in vitro, in contrast to the usual mode of antifungal defense which requires Th17 mediated cell-based responses. These observations suggest that metamorphic XCL1 is capable of a versatile multimodal form of antimicrobial defense.